Rotation Projects at WashU
FIRST-YEAR ROTATION PROJECTS
At least three rotations are required for WashU DBBS students before choosing a thesis lab:
Understanding the intracellular effects of Cationic Amphipathic Drugs on organelles
Rotation at Peterson Lab, 8/2019-11/2019
Brief Description: Cationic Amphipathic Drugs are utilized in various clinical applications, but their underlying intracellular mechanisms are largely unknown. We hypothesized that due to different drug treatments, cell organelles would exhibit different morphology and changes of their abundance, ultimately contributing to distinct mechanisms of cell death. In this rotation, I successfully completed the imaging tests to investigate the intracellular drug effects.
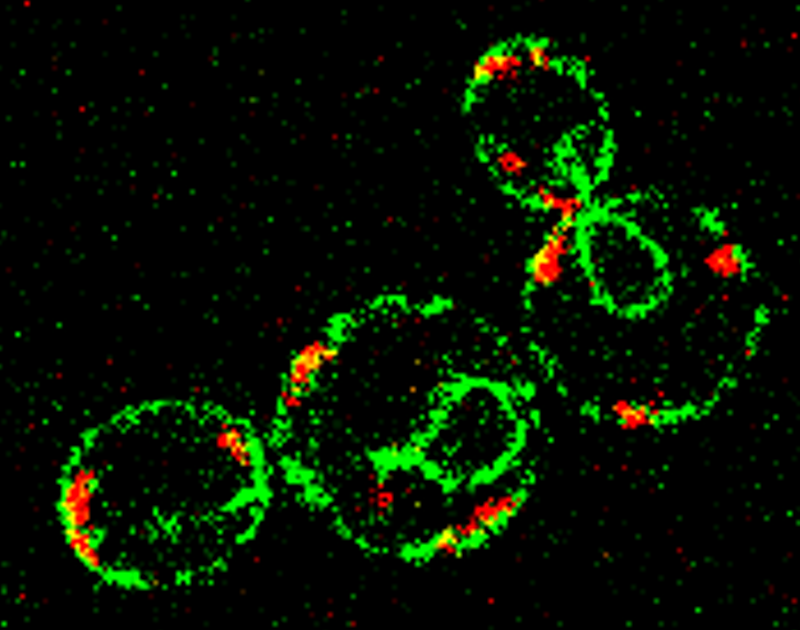
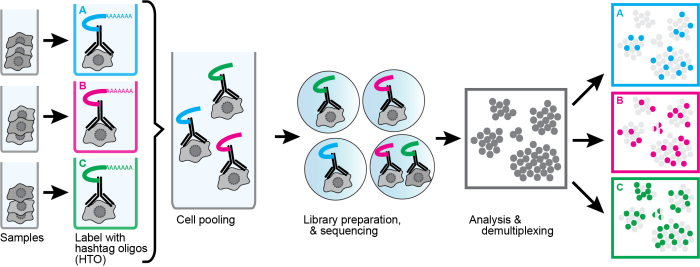
Studying the tumor heterogeneity of head and neck cancer by single-cell sequencing
Rotation at Puram Lab, 11/2019-1/2020
Brief Description: (A) Cell hashing is a single-cell RNA sequencing (scRNA-seq) method multiplexing different cell lines with unique antibody barcodes. It enables us to reduce the routine cost of scRNA-seq, decrease the technology-dependent batch effects, and largely exclude potential doublet signals in scRNA-seq. In this rotation, I successfully performed cell hashing on head and neck squamous cell carcinoma (HNSCC) cell lines which generated a series of datasets with promising data quality.
(B) I also participated another project on functional validations of potential Epithelial–Mesenchymal Transition (EMT)-driving transcriptional factors of HNSCC.
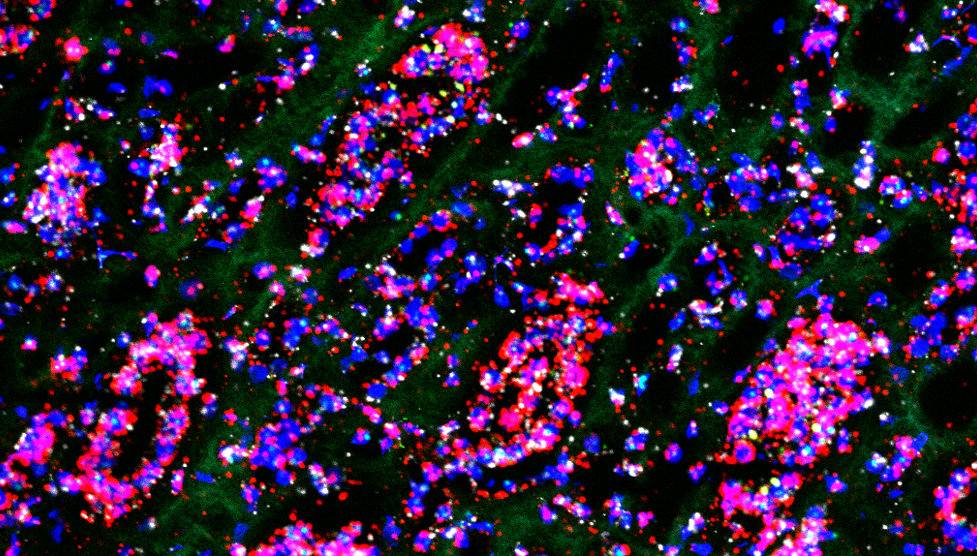
Studying transcriptomic variations in kidney diseases
Rotation at Humphreys Lab, 1/2020-4/2020
Brief Description: (A) RNAscope, a nucleic acid in situ hybridization assay, allows characterization and validation of RNA markers with high sensitivity and specificity. In this rotation, I pioneered the RNAscope assay in the lab on multiple kidney models and successfully identified novel molecular heterogeneity in kidney, as well as transcriptomic variations in certain kidney diseases.
(B) I introduced sci-RNA-seq3 into the lab, which is a cutting-edge high-throughput scRNA-seq technique allowing > 1 million cells being identified per run. sci-RNA-seq3 is expected to establish a comprehensive profile of kidney transcriptomics. This work has been extended to my thesis research.