Graduate (WashU)
WashU DBBS Page
The human kidney consists of ~1 million nephrons, with each nephron composed of ~20 subtypes of glomerular and tubular cells and with diverse stromal and immune populations spanning the organ.
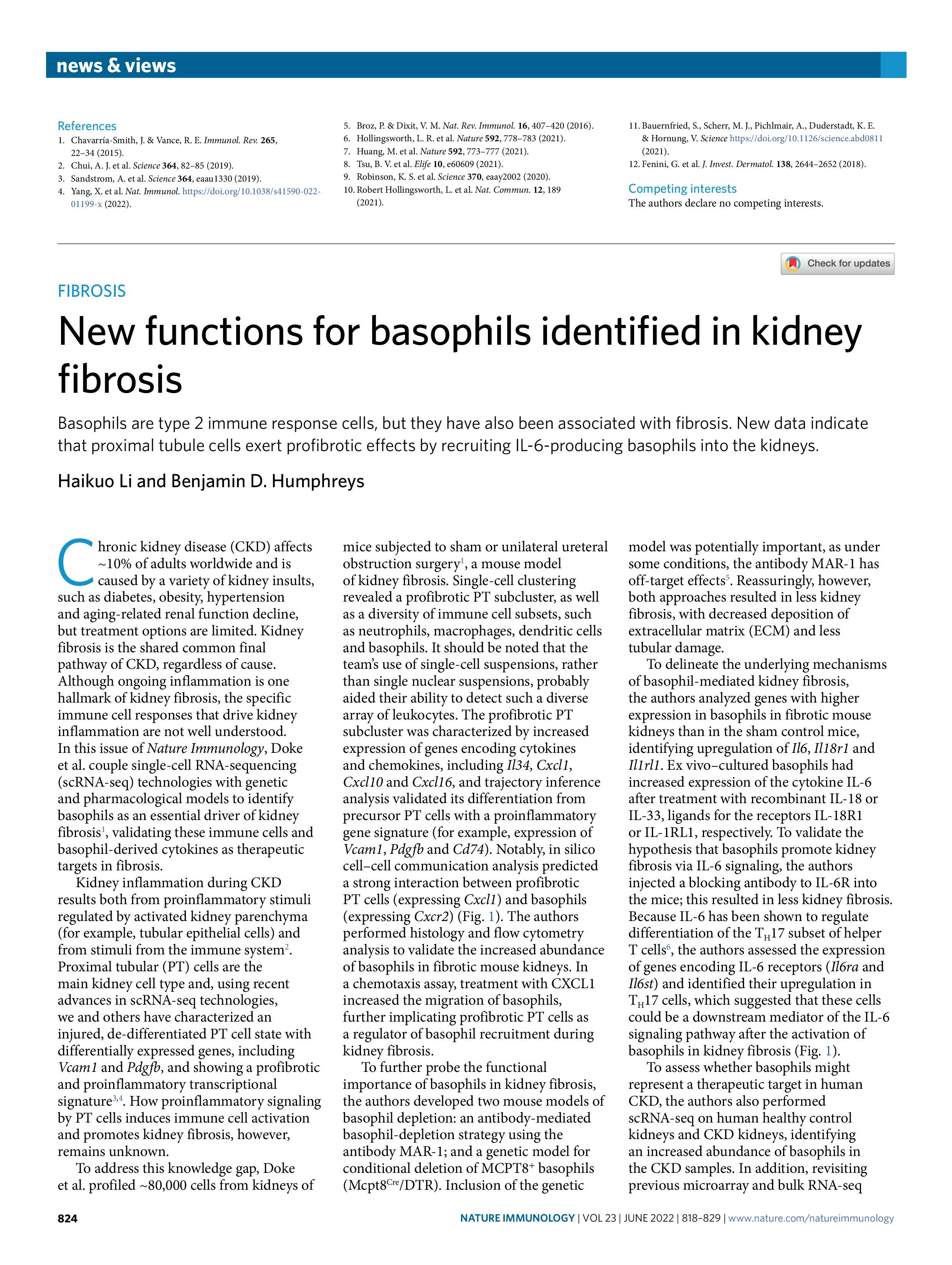
Chronic kidney disease (CKD) affects ~10% of the population worldwide and ultimately can lead to kidney failure. It can be caused by a variety of kidney insults such as diabetes, toxins, hypertension and aging. Kidney fibrosis is a shared final common pathway of all causes of CKD and undermines almost all kidney cell types.
However, the underlying cellular events driving fibrosis are not well understood. For example, injured proximal tubule (PT) cell states exist, but it is unclear whether and how these injured PT cells may drive fibrosis and contribute to disease progression. As another example, lowly abundant cell types (e.g., distal nephron cells) are usually underrepresented by current popular single-cell methods such as 10X Genomics due to low throughput, which limits our understanding of their fibrotic effects and highlights the importance of pursuing large-scale profiling to depict a global view of kidney fibrosis.
My graduate research focuses on investigating molecular mechanisms underlying kidney fibrogenesis with large-scale single-cell multiomics, under the mentorship of Dr. Benjamin Humphreys. With a novel single-cell profiling methodology called split-pool barcoding, we are studying novel cellular states responsible for tissue injury and repair and identifying potential therapeutic targets.
_________________________________________________________________
Our work studying mouse models of kidney fibrosis was published on Cell Metabolism in 2022 (LINK), where we discovered novel injury states of renal proximal tubular cells and defined how lipid metabolism was dysregulated over the time course of disease progression. PLIN2 was identified as a marker of intracellular lipid droplets during kidney injury:
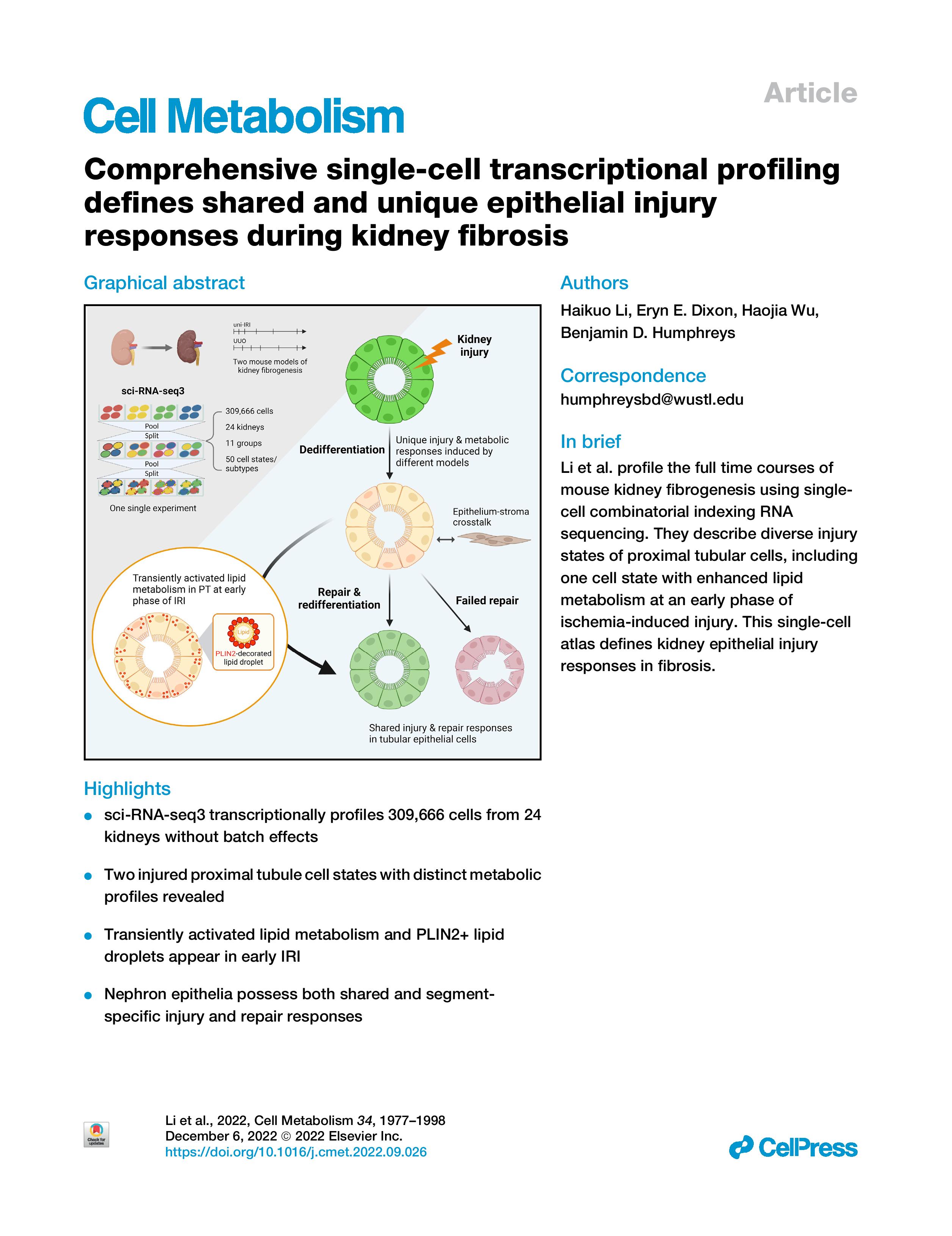 This work was highlighted by NIH NIDDK in the 2024 Annual Report (LINK). The single-cell analysis of proximal tubule injury was selected as the only one scientific illustration in the chapter!
This work was highlighted by NIH NIDDK in the 2024 Annual Report (LINK). The single-cell analysis of proximal tubule injury was selected as the only one scientific illustration in the chapter!

_________________________________________________________________
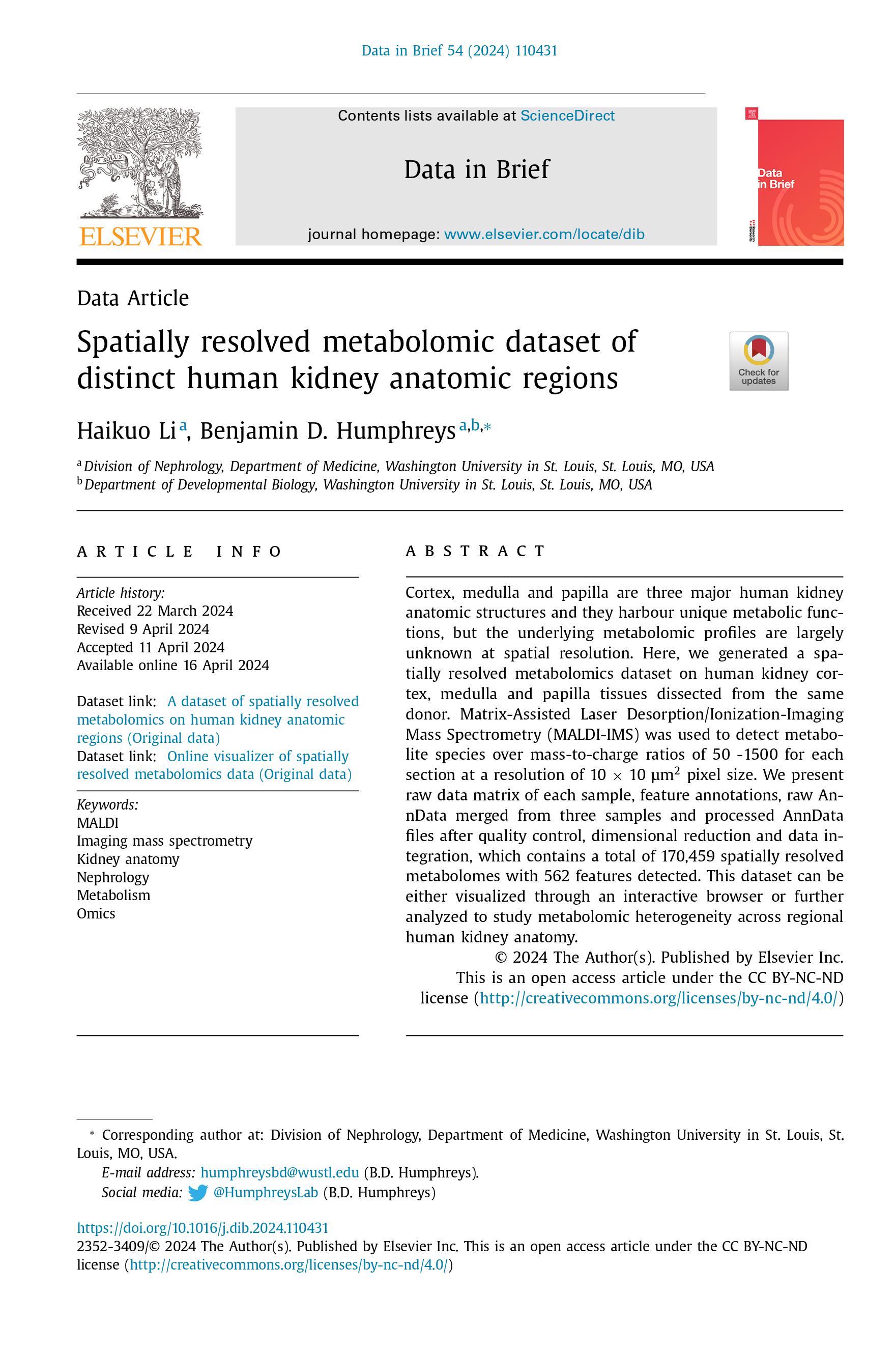
Our work studying human kidney anatomy was published on Cell Metabolism in 2024 (LINK), where we found that the same tubular epithelial cells have distinct signatures depending on regional locations and identified pathogenic candidate genes of human kidney disease. We also presented a computational pipeline for analysis of spatially resolved metabolomics data:
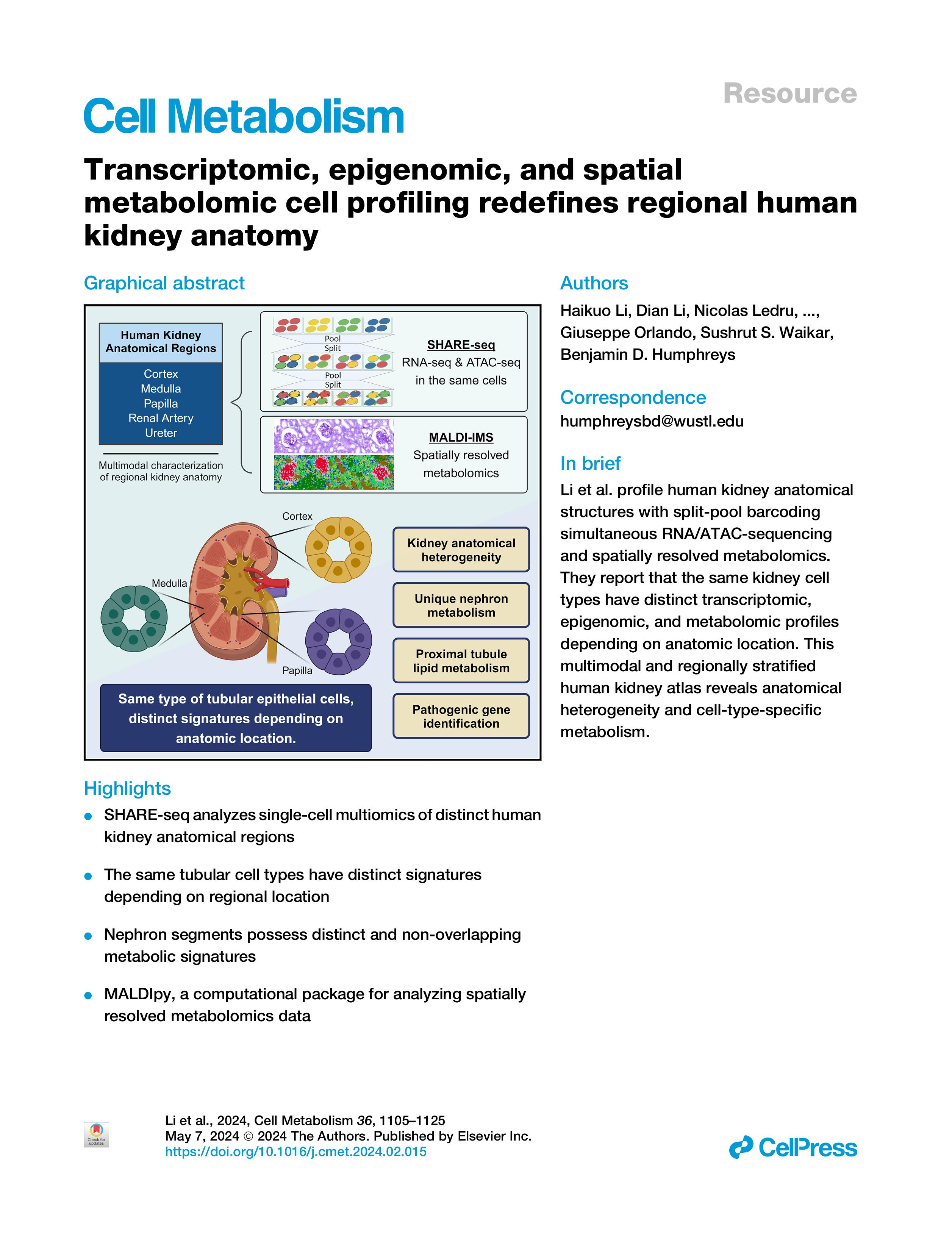
_________________________________________________________________
Our work studying the epigenomic profiles of the human kidney was published on Scientific Data in 2024 (LINK):

_________________________________________________________________
Our optimized sci-RNA-seq3 library generation method with improved library quality on mouse kidney tissues was published on STAR Protocols in 2022 (LINK):
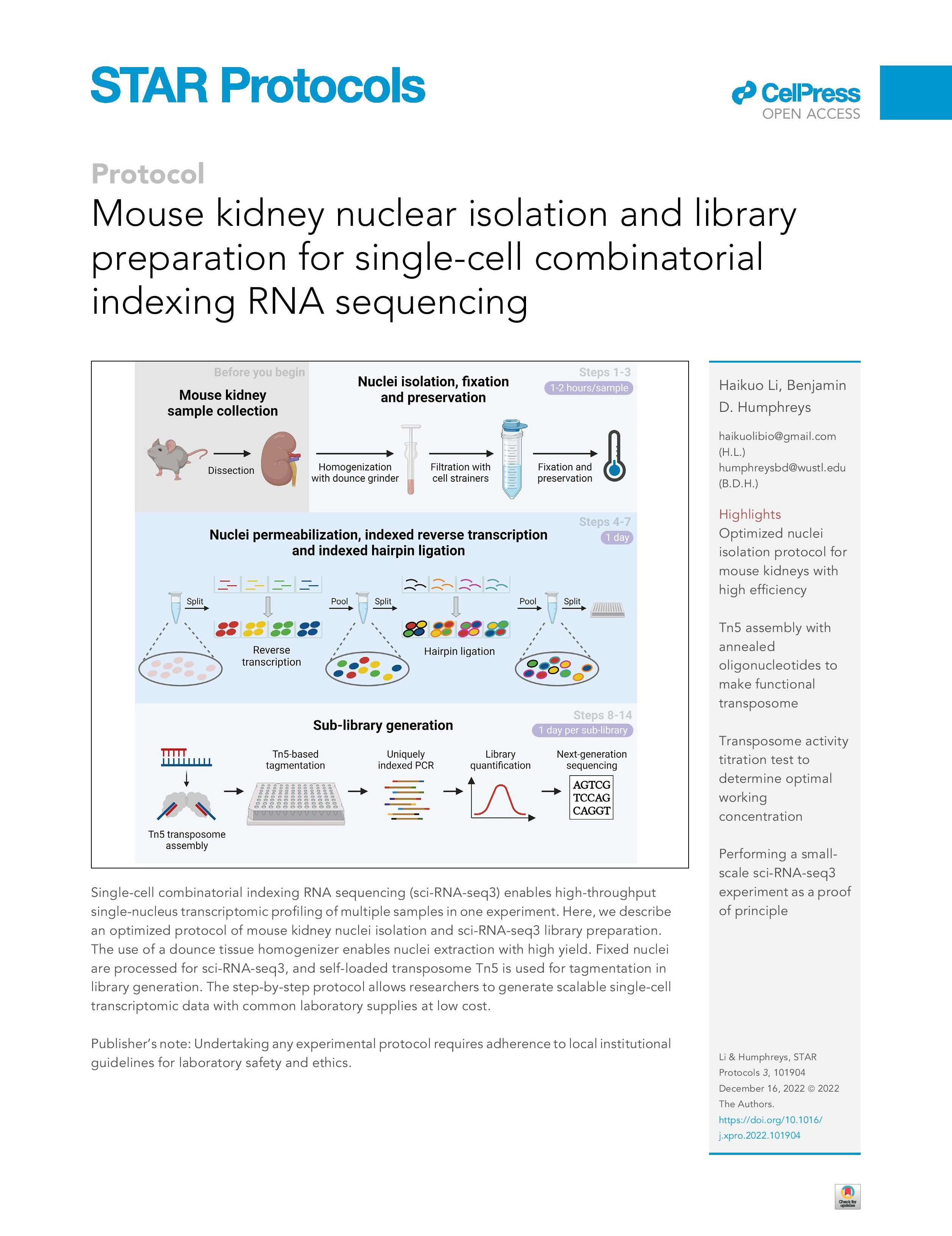
_________________________________________________________________
Our split-pool barcoding multiomics sequencing method, including a protocol for human kidney nuclear isolation, was published on STAR Protocols in 2024 (LINK):
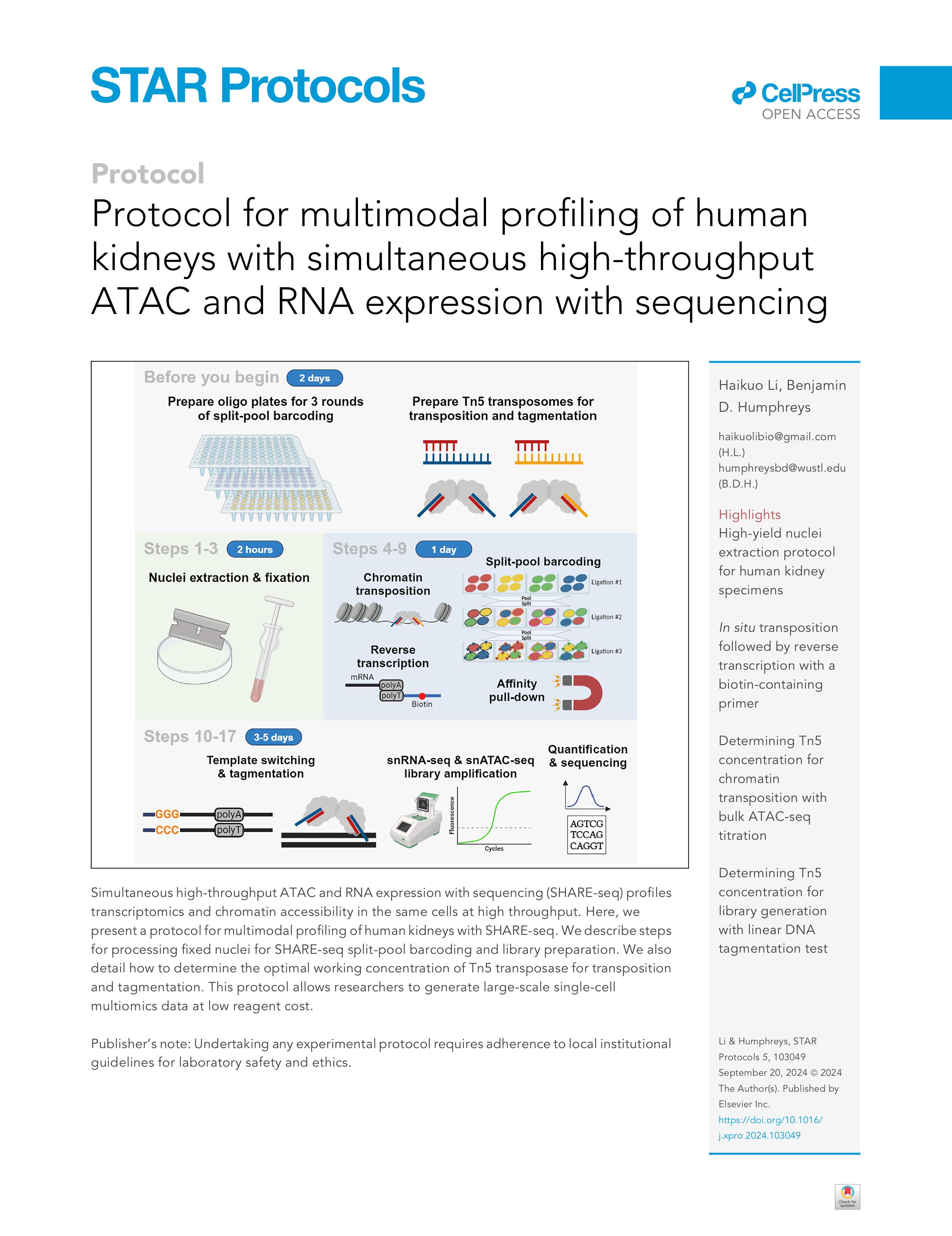
_________________________________________________________________
Our work studying human kidney anatomy with imaging mass spectrometry was published on Data in Brief in 2024 (LINK):
_________________________________________________________________
Glad to chat more about my graduate researches!






_________________________________________________________________
MILESTONES
I was trained by Biochemistry, Biophysics & Structural Biology Program in the first semester in 2019. I developed my research interests in single-cell studies and joined Molecular Genetics and Genomics Program since 2020. I officialy joined Humphreys Lab in June 2020. I passed the qualifying exam in Sept 2020 by proposing a project related to kidney fibrosis and myofibroblast biology. My thesis project was proposed in August 2021. I defended my thesis in December 2023.